4 RStudio and Git/GitHub Setup and Motivation
4.1 Learning Objectives
In this lesson, you will learn:
- What computational reproducibility is and why it is useful
- How version control can increase computational reproducibility
- How to check to make sure your RStudio environment is set up properly for analysis
- How to set up git
4.2 Reproducible Research
Reproducibility is the hallmark of science, which is based on empirical observations coupled with explanatory models. While reproducibility encompasses the full science lifecycle, and includes issues such as methodological consistency and treatment of bias, in this course we will focus on computational reproducibility: the ability to document data, analyses, and models sufficiently for other researchers to be able to understand and ideally re-execute the computations that led to scientific results and conclusions.
4.2.1 What is needed for computational reproducibility?
The first step towards addressing these issues is to be able to evaluate the data, analyses, and models on which conclusions are drawn. Under current practice, this can be difficult because data are typically unavailable, the method sections of papers do not detail the computational approaches used, and analyses and models are often conducted in graphical programs, or, when scripted analyses are employed, the code is not available.
And yet, this is easily remedied. Researchers can achieve computational reproducibility through open science approaches, including straightforward steps for archiving data and code openly along with the scientific workflows describing the provenance of scientific results (e.g., Hampton et al. (2015), Munafò et al. (2017)).
4.2.2 Conceptualizing workflows
Scientific workflows encapsulate all of the steps from data acquisition, cleaning, transformation, integration, analysis, and visualization.
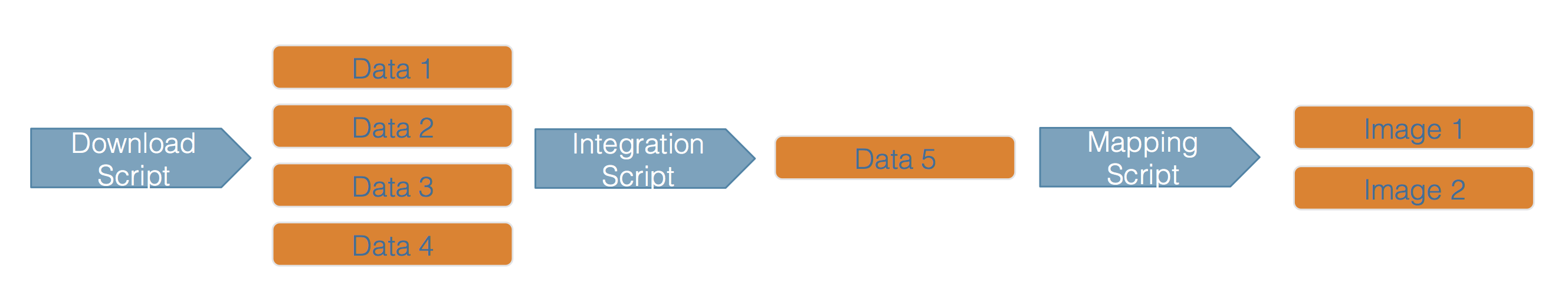
Workflows can range in detail from simple flowcharts to fully executable scripts. R scripts and python scripts are a textual form of a workflow, and when researchers publish specific versions of the scripts and data used in an analysis, it becomes far easier to repeat their computations and understand the provenance of their conclusions.
4.3 Why use git?
4.3.1 The problem with filenames

Every file in the scientific process changes. Manuscripts are edited.
Figures get revised. Code gets fixed when problems are discovered. Data files
get combined together, then errors are fixed, and then they are split and
combined again. In the course of a single analysis, one can expect thousands of
changes to files. And yet, all we use to track this are simplistic filenames.
You might think there is a better way, and you’d be right: version control.
Version control systems help you track all of the changes to your files, without
the spaghetti mess that ensues from simple file renaming. In version control systems
like git, the system tracks not just the name of the file, but also its contents,
so that when contents change, it can tell you which pieces went where. It tracks
which version of a file a new version came from. So its easy to draw a graph
showing all of the versions of a file, like this one:

Version control systems assign an identifier to every version of every file, and
track their relationships. They also allow branches in those versions, and merging
those branches back into the main line of work. They also support having
multiple copies on multiple computers for backup, and for collaboration.
And finally, they let you tag particular versions, such that it is easy to return
to a set of files exactly as they were when you tagged them. For example, the
exact versions of data, code, and narrative that were used when a manuscript was
submitted might be R2 in the graph above.
4.4 Checking the RStudio environment
4.4.1 R Version
We will use R version 3.6.2, which you can download and install from CRAN. To check your version, run this in your RStudio console:
If you have R version 3.6.1 that will likely work fine as well.
4.4.2 RStudio Version
We will be using RStudio version 1.2.500 or later, which you can download and install here To check your RStudio version, run the following in your RStudio console:
If the output of this does not say 1.2.500 or higher, you should update your RStudio. Do this by selecting Help -> Check for Updates and follow the prompts.
4.4.3 Package installation
Run the following lines to check that all of the packages we need for the training are installed on your computer.
packages <- c("dplyr", "tidyr", "devtools", "usethis", "roxygen2", "leaflet", "ggplot2", "DT", "scales", "shiny", "sf", "ggmap", "broom", "captioner", "MASS", "doParallel", "foreach", "parallel")
for (package in packages) { if (!(package %in% installed.packages())) { install.packages(package) } }
rm(packages) #remove variable from workspace
# Now upgrade any out-of-date packages
update.packages(ask=FALSE)If you haven’t installed all of the packages, this will automatically start installing them. If they are installed, it won’t do anything.
Next, create a new R Markdown (File -> New File -> R Markdown). If you have never made an R Markdown document before, a dialog box will pop up asking if you wish to install the required packages. Click yes.
At this point, RStudio and R should be all set up.
4.5 Setting up git
If you haven’t already, go to github.com and create an account. If you haven’t downloaded git already, you can download it here.
Before using git, you need to tell it who you are, also known as setting the global options. The only way to do this is through the command line. Newer versions of RStudio have a nice feature where you can open a terminal window in your RStudio session. Do this by selecting Tools -> Terminal -> New Terminal.
A terminal tab should now be open where your console usually is.
To see if you aleady have your name and email options set, use this command from the terminal:
To set the global options, type the following into the command prompt, with your actual name, and press enter:
Next, enter the following line, with the email address you used when you created your account on github.com:
Note that these lines need to be run one at a time.
Finally, check to make sure everything looks correct by entering these commands, which will return the options that you have set.
4.5.1 Note for Windows Users
If you get “command not found” (or similar) when you try these steps through the RStudio terminal tab, you may need to set the type of terminal that gets launched by RStudio. Under some git install senerios, the git executable may not be available to the default terminal type. Follow the instructions on the RStudio site for Windows specific terminal options. In particular, you should choose “New Terminals open with Git Bash” in the Terminal options (Tools->Global Options->Terminal).
In addition, some versions of windows have difficults with the command line if you are using an account name with spaces in it (such as “Matt Jones”, rather than something like “mbjones”). You may need to use an account name without spaces.
4.6 Updating a previous R installation
This is useful for users who already have R with some packages installed and need to upgrade R, but don’t want to lose packages. If you have never installed R or any R packages before, you can skip this section.
If you already have R installed, but need to update, and don’t want to lose your packages, these two R functions can help you. The first will save all of your packages to a file. The second loads the packages from the file and installs packages that are missing.
Save this script to a file (eg package_update.R).
#' Save R packages to a file. Useful when updating R version
#'
#' @param path path to rda file to save packages to. eg: installed_old.rda
save_packages <- function(path){
tmp <- installed.packages()
installedpkgs <- as.vector(tmp[is.na(tmp[,"Priority"]), 1])
save(installedpkgs, file = path)
}
#' Update packages from a file. Useful when updating R version
#'
#' @param path path to rda file where packages were saved
update_packages <- function(path){
tmp <- new.env()
installedpkgs <- load(file = path, envir = tmp)
installedpkgs <- tmp[[ls(tmp)[1]]]
tmp <- installed.packages()
installedpkgs.new <- as.vector(tmp[is.na(tmp[,"Priority"]), 1])
missing <- setdiff(installedpkgs, installedpkgs.new)
install.packages(missing)
update.packages(ask=FALSE)
}Source the file that you saved above (eg: source(package_update.R)). Then, run the save_packages function.
Then quit R, go to CRAN, and install the latest version of R.
Source the R script that you saved above again (eg: source(package_update.R)), and then run:
This should install all of your R packages that you had before you upgraded.
References
Hampton, Stephanie E, Sean Anderson, Sarah C Bagby, Corinna Gries, Xueying Han, Edmund Hart, Matthew B Jones, et al. 2015. “The Tao of Open Science for Ecology.” Ecosphere 6 (July). https://doi.org/http://dx.doi.org/10.1890/ES14-00402.1.
Munafò, Marcus R., Brian A. Nosek, Dorothy V. M. Bishop, Katherine S. Button, Christopher D. Chambers, Nathalie Percie du Sert, Uri Simonsohn, Eric-Jan Wagenmakers, Jennifer J. Ware, and John P. A. Ioannidis. 2017. “A Manifesto for Reproducible Science.” Nature Human Behaviour 1 (1): 0021. https://doi.org/10.1038/s41562-016-0021.